Constrained non-negative matrix factorization for calcium imaging data analysis
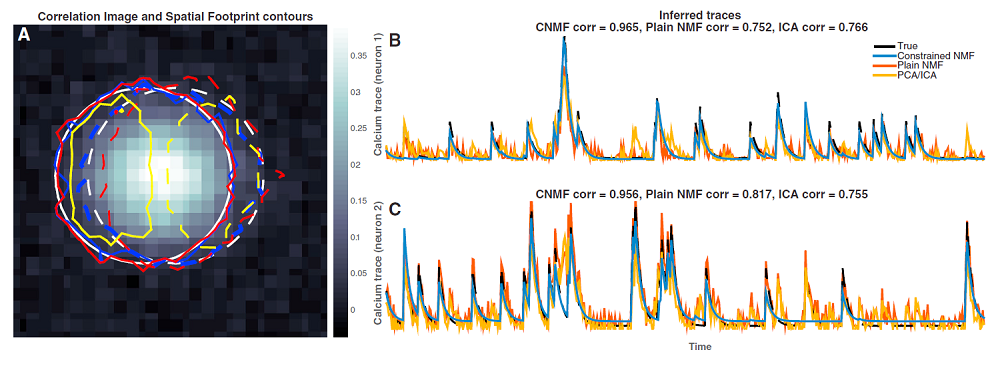
I tweeted about this last fall. This is the best algorithm I’ve seen for segmenting and extracting time course from calcium imaging data. Eftychios Pnevmatikakis developed the code in Liam Paninski’s lab. The work is reported in a pair of papers in Neuron, and the code is freely available (links below).
The source separation works so well that Darcy Peterka used it to demix imaging data where two planes were imaged simultaneously. This isn’t temporal multiplexing (as we’ve been doing in my lab; Stirman et al.), it’s complete crosstalk between two imaging planes. They (Yang et al.) used an SLM to create two focii which were simultaneously scanned over two imaging planes (displaced in Z and/or XY). They were able to completely demix the data and it looks very good. This is an interesting way to increase data acquisition throughput.
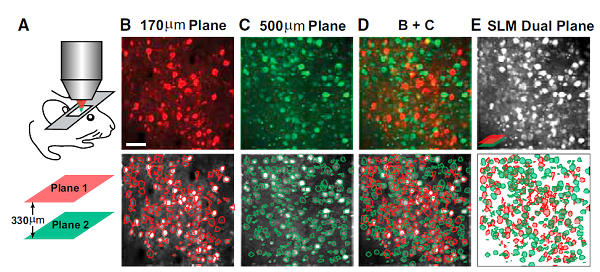
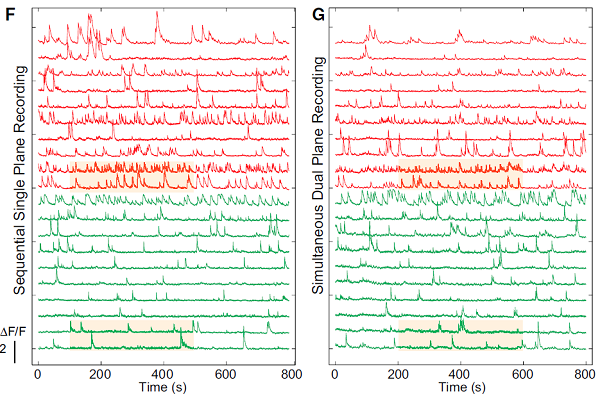
These are very exciting advances. We’re already using them in my own lab. Eftychios’ code basically works out-of-the-box. We’ve had to do a bit of tweaking to the algorithms depending on the attributes of particular data sets, but that’s to be expected. Eftychios has been very, very helpful (thanks!).
So try these things out and give them feedback!
Pnevmatikakis et al. (Algorithm, performance on example applications)
Yang et al. (Dual-plane imaging)
Github page with code
It’s also available for Python:
https://github.com/agiovann/Constrained_NMF
Some initial testing suggests that on the same machine, the Python version is a bit slower but can handle larger data sets without running out of memory.
[…] data. It is a nice quick read and discusses earlier work in the area. (That’s Eftychios of constrained-non-negative-matrix-factorization-for-calcium-imaging-analysis […]