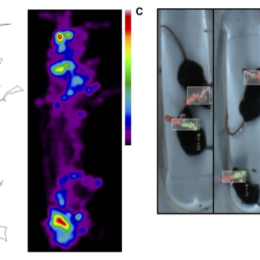
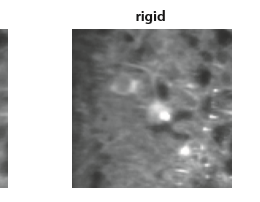
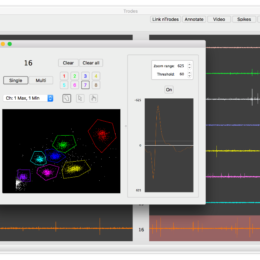
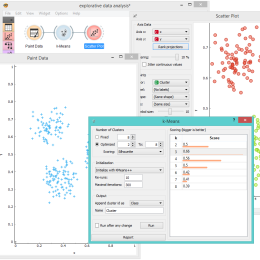
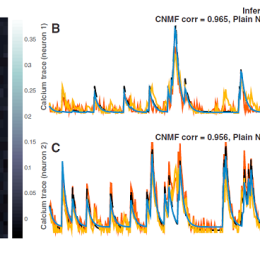
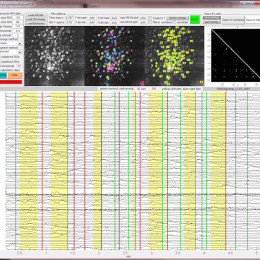
Server space – cloud or on-campus?
Cloud storage is not the cure-all it is sometimes proposed to be. On-campus servers have some advantages in transmission speed and cost, especially as storage requirements grow considerably. Talk to your campus IT services before shelling…