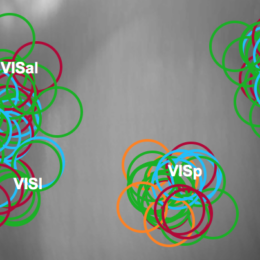
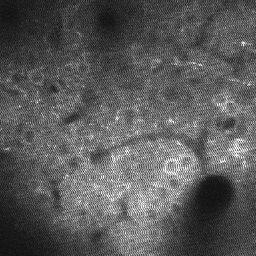
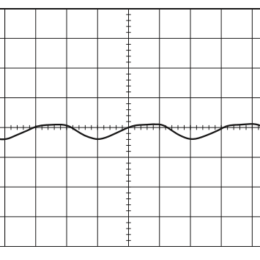
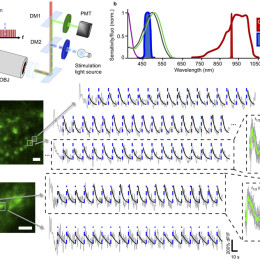
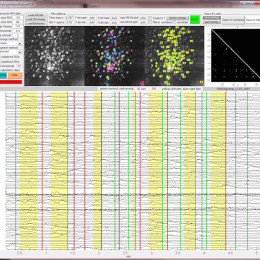
Open, easy laser-scanning microscope software: LSMAQ
Years ago, Benjamin Judkewitz wanted to try some new techniques out with laser scanning two-photon microscopy. However, the software we were using was rather cumbersome to modify. So he wrote his own software from scratch…