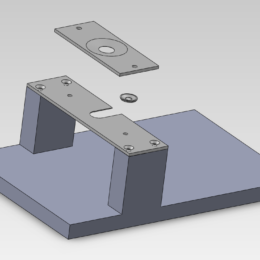
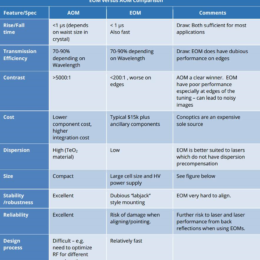
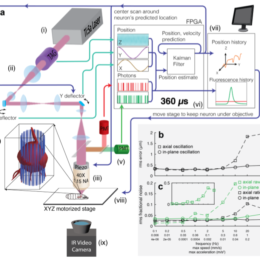
Guidance for designing an objective from scratch
Several people have reached out for guidance on designing and building an objective from scratch, when they have little-to-no background in custom optics. Here are some notes on the topic, in broad strokes, focused on the practicalities.
…