HDF5 + XML = SDCubes
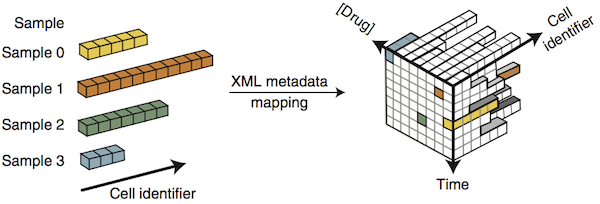
A paper on data organization just came out in Nature Methods (Millard et al. 2011, commentary by Swedlow et al.). They believe, as do I, that using XML schema to organize data is a good way to simplify automated analysis. For the primary container, they use HDF5. Fun fact for all the MATLAB users in the audience (from Tim at Imperial College):
The .mat file format is simply an HDF5 file with a pointless header prepended.
They call the XML metadata+HDF5 data combos “SDCubes”, for Semantically typed Data hyperCubes. Why cubes? That suggests that they are the same size along all axes, which they probably aren’t. If you don’t like hyperrectangle, you can use orthotope. One point that is lost in the figure I put above is the idea that the axes do not have to be continuous. There can be gaps and jumps. There can also be piles of data that all share one point on an axis, if that suits the data.
I like this approach because it is very general and simple. It consists of two file formats that are already being used by many researchers. In a way, the authors didn’t “create” anything. Hopefully this paper will give the strategy some added credibility and help to standardize it. Then people can concentrate on developing tools for working with data in this system, rather than developing new formats all the time.
Hi Spencer,
I like SDCube as well. I wrote about it and some related approaches here: http://digitheadslabnotebook.blogspot.com/2011/12/sdcube-and-hybrid-data-storage.html
We’ve struggled to wedge scientific data into relational databases and it works, but it seems to fight you every step of the way. The strategy of using data stores more naturally suited to particular data-types makes a lot of sense – XML for hierarchical data, HDF for numeric matrices. Networks are another data structure that fit only unwillingly into SQL DBs.
– Chris
[…] Graupner has coded a nice program for browsing and managing hdf5 files (closely related to MATLAB files) called hdf5Manager. And it’s open […]