Analysis algorithms: performance quantification and ground truth
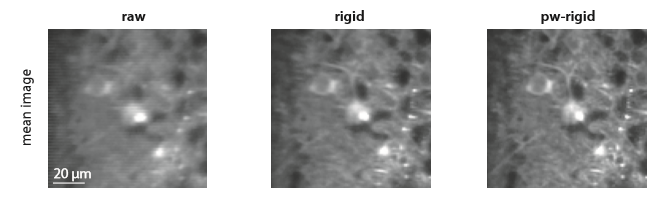
We recently tweeted about a preprint from Eftychios A Pnevmatikakis and Andrea Giovannucci (code). The preprint is on motion correction for calcium imaging data. It is a nice quick read and discusses earlier work in the area. (That’s Eftychios of constrained-non-negative-matrix-factorization-for-calcium-imaging-analysis fame).
Marius Pachitariu recognized the algorithm as very similar to one that he uses in his own Suite2p software (preprint, code; which is excellent by the way, and we highly recommend trying it out). He initially indicated that the authors were “exactly copying” his Suite2p algorithm, but later clarified that the algorithms are at least somewhat different. Indeed, in the first table in the preprint, they compare their algorithm to the Suite2p algorithm and get quantitatively different results, so it’s not an exact copy. The differences between the algorithms may be small, but small differences can matter. For example, in work on convolutional neural networks, it was found that rectified linear units provide much faster learning than tanh or sigmoid functions (Dahl et al., Krizhevsky et al.). The overall network architecture was the same, they just changed the activation function, and found useful effects on performance. So while we certainly appreciate Marius’s insight into the algorithm, similarity to previous work does not diminish our interest in Pnevmatikakis & Giovannucci’s work.
Postscript
While quantitative comparison of algorithm performance is straightforward, it is often performed using “fake” data, and so it doesn’t tell us how close we get to the truth. Measuring performance on fake data can determine whether two algorithms are different, but it cannot tell use which one is better. Marius rightly pointed out that it would be best to have ground truth data for such evaluations. Unfortunately, that’s not easy to come by– see the discussion below, and this recent review.
With the rapid adoption and growth in the use of large-scale neuronal sampling (be it from electrode arrays or calcium imaging), there has been pressure to relax the methodological rigor. Sometimes this is for practical reasons. For example, it’s technically very difficult to obtain simultaneous electrophysiology data when imaging through GRIN lenses because the working distance is so small. Even in more accessible systems where simultaneous electrophysiology is possible, the nonlinear and highly complex relationships between spiking activity and fluorescence signals from genetically encoded calcium indicators are not routinely measured, and when they are, they are only roughly approximated. Often, more complete characterization may not yield precision that would inform a particular study’s results. Still, we shouldn’t ignore the relaxed rigor, because doing so would certainly eventually come back to haunt us.
Fortunately, many in our community care dearly about the rigor with which these experiments are performed, and are working collaboratively to test algorithms against ground truth data sets. For example, the NeuroFinder Challenge is an important effort in this area (Marius is doing quite well in that, by the way).
Check the preprint comments for further discussion by the authors:
http://biorxiv.org/content/early/2017/02/14/108514