An Arch-based voltage sensor
The newest voltage sensor is from Adam Cohen’s lab and based on a microbial rhodopsin, a class of light sensitive proteins already being put to use in optogenetics.
In this case, they’ve take Archaerhodopsin 3 (a related protein to Arch, which has been used to hyperpolarize neurons and silence activity) and removed it’s ion fluxing ability. But since it still senses voltage and light, its optical characteristics change with voltage.
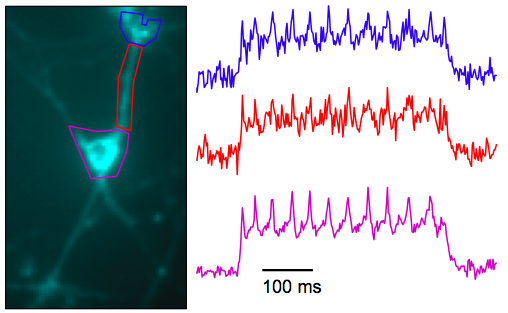
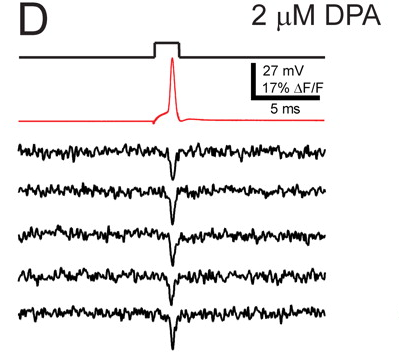
How does it compare with existing voltage sensors? It has a relatively large change in fluorescence over a physiological voltage range (about 50%), but it’s very slow (41 ms onset). They can nicely see APs in single sweeps in cultured cells. In slices and in vivo, the signals will be smaller. One of the best voltage imaging schemes around, DiO+DPA, has a similar fluorescence change and is way faster (<1 ms). Note that signals from Arch(D95N) in culture (top picture below) don’t look massively better than DiO+DPA in slice (bottom picture below). Hopefully Arch(D95N)’s signals will still be usable in slices and in vivo.
Arch(D95N) in culture
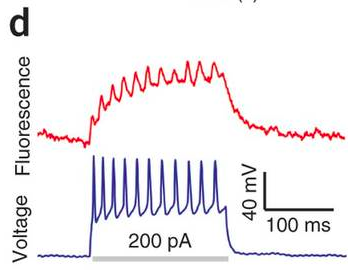
DiO+DPA in slice
But of course, having a genetically targettable construct is a huge advantage. So a fairer comparison would be to other voltage sensing proteins. In that case, Arch(D95N)’s fluorescence change is excellent, but its slow speed is a handicap and makes that big fluorescence change less useful. Perhaps the biggest problem with Arch(D95N) is that it’s dim. The quantum yield is 9e-4 and an EMCCD was required for these experiments. For comparison, many GFP based proteins have quantum yields over 5e-1. It’s extiction coefficient is decent– it absorbs light– it’s just really unlikely to emit a fluorescence photon.
Maybe the protein engineers will follow up and make a brighter and faster version, the authors themselves report that they’re actively working on speed. For what it’s worth, the native protein (with it’s fluxing capability intact) is much faster, so there’s hope on that front.
Knopfel has taken perhaps the most rigorous and deliberate approach to voltage sensing protein design. The first big problem is that the sensor has to be at the membrane, so unlike calcium sensors, the signal-generating ROI is pretty small. The second big problem is that the sensor adds an unnatural capacitance to the membrane, so when expression levels increase to improve S:N, the capacitance also increases and messes up excitability. They have a nice analysis in this paper.
Nevertheless, there’s a lot to do with voltage sensors and I don’t think people have really started to fully explore what they’re capable of. These systems don’t need to be perfect, there is a huge amount of physiology to explore even using the existing voltage sensors.