GCaMP5: Already old news?
The GCaMP5 paper is out now. The paper looks fantastic. There’s tons of helpful information and impressive demonstrations of its performance in several popular model systems.
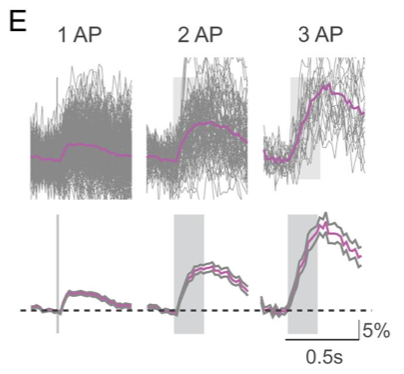
But isn’t GCaMP6 the best thing going? Is this old news? GCaMP6 detects single APs better than OGB-1 (which, as the top figure shows, GCaMP5 does not). This is what the same group is presenting at SfN (emphasis mine):
927.08/FFF77
Reliable detection of single action potentials and synaptic calcium signals using improved genetically-encoded calcium indicators
*T.-W. CHEN, J. YU, R. A. KERR, V. JAYARAMAN, L. L. LOOGER, K. SVOBODA, D. S. KIM;
HHMI Janelia Farm Res. Campus, Ashburn, VA
Genetically-encoded calcium indicators (GECIs) allow non-invasive measurement of neuronal activity in vivo. Currently available GECIs, such as the widely used GCaMP3, still suffer from limited sensitivity. Using structure-guided mutagenesis and high-throughput screening, we increased the fluorescence change in response to single action potentials (APs) by >10-fold compared to GCaMP3. We also accelerated the kinetics by ~2-fold. These new GECIs reliably report single APs in single trials in vivo with near 100% accuracy. In the mouse visual cortex, we detected ~5-fold more visually responsive neurons. The sensitivity, dynamic range and speed of the new GECIs exceed those of the synthetic indicator OGB-1. The improved sensitivity further facilitated reliable measurement of synaptic calcium signals in the dendrites of pyramidal cells and parvabumin (PV)-positive interneurons in vivo. Hot spots of orientation-selective domains can be resolved both in single pyramidal cell spines and small segments of PV cell dendrites. These improved GECIs will permit a more complete description of neuronal circuit function and enable long-term functional imaging of single synapses.
207.14/DDD38
Engineering next generation GCaMP calcium indicators using neuron-based screening
T.-W. CHEN-1, T. J. WARDILL-2, J. P. HASSEMAN1, G. TSEGAYE1, B. F. FOSQUE-1, E. R. SCHREITER-1, B. E. KIMMEL-1, R. A. KERR-1, V. JAYARAMAN-1, K. SVOBODA-1, L. L. LOOGER-1, *D. S. KIM-1;
1-Janelia Farm Res. Campus, Howard Hughes Med. Inst., Ashburn, VA; 2-Marine Biol. Lab., Woods Hole, MA
Neuronal activity can be imaged in vivo using GCaMP, a green fluorescent, genetically-encoded calcium indicator. GCaMP and its variants detect calcium concentration changes in neurons, which can be used as a measure of electrical activity. Hundreds of neurons can be monitored simultaneously and non-invasively, and activity of distinct neurons can be followed using specific gene regulatory elements. However, current GCaMP variants in widespread use exhibit relatively low sensitivity and slow kinetics. To improve GCaMP performance, we created libraries of novel variants by structure-guided, site-directed mutagenesis and assayed activity using neuron-based screening. Both the amplitude and kinetics of neuronal calcium concentration changes are difficult to mimic in purified protein assays or non-neuronal cells. We therefore developed a neuron-based screen. GCaMP variants were tested in cultured rat primary hippocampal neurons transduced by lentiviral infection. Action potentials were stimulated using field electrodes, and fluorescence imaging was performed on a motorized microscope in an automated manner. We report higher sensitivity GCaMP variants that can reliably detect single action potentials in neurons and variants with faster kinetics. The highest sensitivity variant exhibited a 10-fold greater single action potential response compared with GCaMP3 and exceeded the response of the synthetic dye, Oregon Green BAPTA-1 (OGB-1). Even though high sensitivity variants generally exhibited relatively slower kinetics, some high sensitivity/high speed variants were identified. One variant was twice as fast as GCaMP3 in terms of decay kinetics, with 7-fold improved sensitivity. The rise kinetics were also faster. These variants will be especially useful for measurement of high frequency neuronal activity and action potential timing. Increased sensitivity in the neuronal assay was correlated with improved calcium affinity using purified protein, but kinetic parameters were not predictable. These next generation GCaMPs will enable improved correlation of activity patterns of large ensembles of neurons in vivo with animal behavior.
Andrew Hires has a post up on Brainwindows on the paper and its publication. He writes, “…the paper was repeatedly delayed. Why? Because reviewers viewed it as ‘too incremental’ of an upgrade, and not worth publishing in a prominent journal (no, I’m not talking Nature or Science level).” He doesn’t say what journal(s) delayed it, but there aren’t very many journals between Nature/Science and J Neurosci. To me, the key issue is this, again quoting from Andrew’s post: “Multiple post-docs spent years of their lives developing and carefully testing this tool. They deserve a quality publication for their efforts.”
Tool development is as tough as any other type of science. However, unlike research findings that retain their valuable novelty over the years, it’s typically only a matter of time before an improved version of a tool is developed. Then reports of earlier versions of that tool are less likely to be cited. But even so, the earlier tools are often widely implemented for at least a brief period of time, and enable new experiments and new science.
I think improved GECIs, voltage sensors, optogenetic constructs, and other similar tools should receive special considerations by journals for two reasons: (1) they are of unusually broad interest, and (2) their relevancy is fleeting. The nature of the science is a bit different, and so the resulting report and review process should be a bit different as well.
Dear Editors– Instead of insisting on encyclopedic demonstrations of an improved tool’s quantitative performance across the entire animal kingdom of model systems through a dizzying consortium of heavyweight labs; perhaps put some weight on the endorsements of prominent early adopters, streamline the process, and let the tool makers get back to what they do best: inventing the next best thing.
as a tech developer of over 20 yrs I have to say this is the issue we face!
one man’s increment is another’s giant leap.
there is no simple answer to the “top journal” problem.
My old boss (Jack Kaplan) said to me yrs ago: There are no good journals, only good papers.
this full paper looks great for being published as such a full study.
Best wishes to your new lab!
Graham Ellis-Davies
Grab it while it’s still hot! No time to waste – GCaMP-whatever is certainly right around the corner!
GCaMP6f,m,s available for preorder on PennU in all its Syn/Cag/FLEX AAV glory:
http://www.med.upenn.edu/gtp/vectorcore/WhatsNew.shtml#GCaMP6
And ready to order on addgene:
http://www.addgene.org/browse/article/5916/
Cheers,
T