DIY Superresolution, part II
This paper (York et al. 2012) came out earlier this year, but I thought it’s worth highlighting here, give the subject matter.
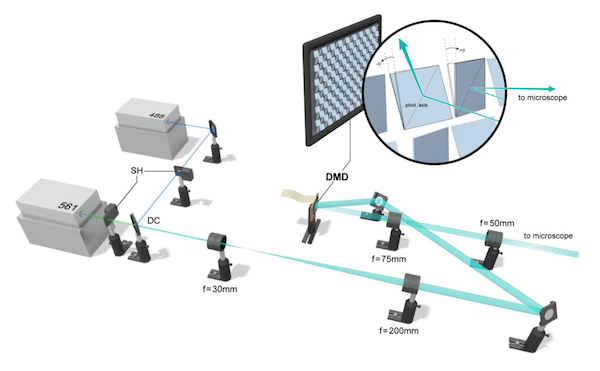
We present a hybrid technique, multifocal SIM (MSIM), that combines the resolution-doubling characteristics of SIM with the physical optical sectioning of confocal microscopy. MSIM uses sparse 2D excitation patterns generated with a DMD integrated into a conventional wide-field microscope and digital processing after acquisition (‘postprocessing’) to obtain optically sectioned images with ~145-nm lateral and ~400-nm axial resolution at 1-Hz frame rates. Relative to existing SIM, our implementation is easier to integrate onto existing microscopes and is considerably cheaper than commercial SIM.
For imaging, they used an sCMOS camera: the pco.edge (I’ve demo’d one, they’re very cool). In the supplement, they offer some helpful tips on how to set up the camera to obtain high quality images. This section is a good example of how the authors carefully prepared the paper to ensure that others can replicate their success– all methods papers should be this detailed and helpful.
The lead author, York, recently commented in the confocal listserv and shared some of his thoughts and code on the pco.edge camera (link).
I wish I could build a confocal/MP microscope by myself. As for the superresolution microscopy, I am not impressed at all with results shown in articles. If superresolution microscopy is to be useful, they should at least 1) image some subcellular structures on their own without using prior information from electron microscopy and 2) able to identify protein interaction. I frequently check superresolution papers published both by big shots and newbies and found that they are just showing images of structures easily resolvable with confocal microscope. Many show images comparing wide-field with superresolution instead of comparing confocal with superresolution microscopy.
I’ve seen in some papers that the authors compare the superresolution images with deconvolved confocal images (sometimes in the supplement– perhaps in response to skeptical reviewers?). Often the differences are quite small. That said, it *is* a real, quantitative increase in resolution. And although I share your desire to see more biological findings using superresolution imaging, perhaps it will simply take time for the technology to make a real impact. Users will need to figure out the strengths and weaknesses and how to design their experiments to capitalize on the technology.
For example, one basic neuroscience application for which superresolution could be useful for is identifying whether a tagged protein is pre- or post-synaptic. Confocal resolution is just at the cusp of being able to determine this, but superresolution images should offer definitive evidence.
What applications do you think superresolution imaging might be useful for? What’s some of the best biology that has been done with it so far?
[…] for excellent Labrigger community members like htwe, they include comparisons to confocal imaging, rather than widefield […]
Hello, this is Andrew, an author of the paper discussed here. Glad to see interest in the technique. The comments raise some interesting points that I think are worth further discussion. I’ll start with Htwe:
“””
1) image some subcellular structures on their own without using prior information from electron microscopy
“””
I assume you’re referring to using microtubules and beads as a resolution target. (We certainly don’t use prior information about structure in any of our processing.)
There’s an interesting tension here. Imaging well-known objects (like microtubules) tests the microscope and helps rule out artifacts, but it’s boring and you learn no biology. Imaging unknown objects tests biology, but if you see anything that’s never been seen before, you must ensure it’s not an artifact. Of course the best approach is to do both – validate the scope, then go exploring.
We used microtubules for validation, and histones in worms for exploring. We did see some sub-nuclear structure in worm histones (see Supp. Fig. 10 in our paper). Our biologist collaborators had the correct reaction – interested but skeptical. They don’t see these structures in their spinning disk microscope, so how do we know if they’re real? In this case, we were able to observe the same structure with several unrelated microscopy techniques, and this convinced them (and us) they’re real. This type of question will come up again I’m sure, and it will always be fun figuring out the appropriate sanity checks.
“””
2) able to identify protein interaction
“””
We took some 2-color timelapses of cell-in-gels. However, I’m not a biologist, so there’s a good chance I’m misunderstanding your suggestion. What types of targets would you find most interesting for a new device to demonstrate utility?
Our microscope development cycle is idea-centered rather than goal centered, so I build a device with new capabilities first and recruit biologist collaborators second. Matching interesting biological questions to new capabilities is a bit of an art, especially since most biologists shape their questions to suit existing capability.
“””
…just showing images of structures easily resolvable with confocal microscope
“””
Excellent point! Resolution doubling is nice, but it’s a rare biology question which is answered by 150 nm resolution that is unanswerable with 300 nm resolution. However, such problems do exist! For a nice example of such a problem, check out a recent paper by Eswaramoorthy et al: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3225972/
“””
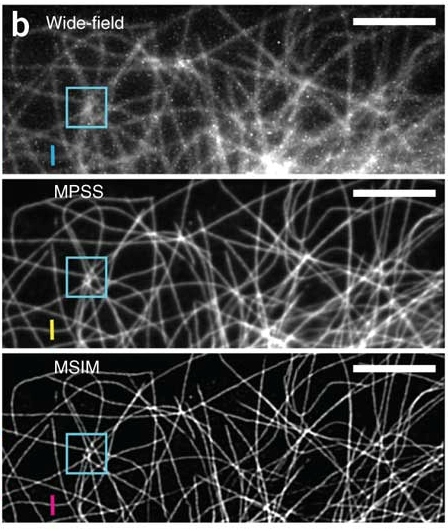
Many show images comparing wide-field with superresolution instead of comparing confocal with superresolution microscopy.
“””
Very true. I think this is because Mats Gustafsson did this in his first SIM paper, and the community is following his lead. I think he compared to widefield in order to establish his “resolution doubling” claim, which was extraordinary at the time.
We compare to both widefield (Fig 1) and confocal (Supp. Fig. 9). As with any microscope comparison, a truly fair comparison is complicated, but we did our best. In our hands, MSIM gave better pictures than a point-scanning confocal (higher resolution, higher SNR, similar acquisition speed). Is this generally true? Only one way to find out! We’ve turned the microscope over to the NIH community, and we’ll see if it produces any useful insight for biology.
“””
…it will simply take time for the technology to make a real impact. Users will need to figure out the strengths and weaknesses and how to design their experiments to capitalize on the technology.
“””
I couldn’t agree more! Especially since the advantages of new technology are often offset by tradeoffs. In our case, processing time and user interface are worse than for a commercial point-scanning confocal, and our acquisition speed is worse than a spinning-disk.
We work with many biologists who are expert microscopists and understand why their work will benefit from our new capability, but not every collaborator who comes to us actually needs higher resolution or better SNR.