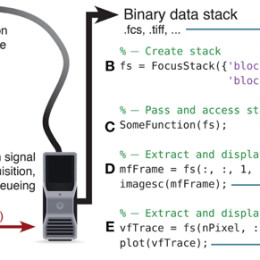
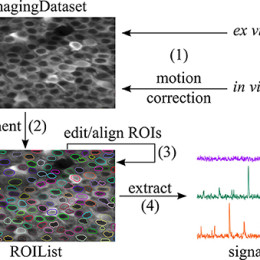
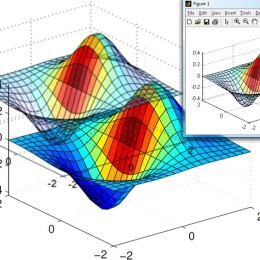
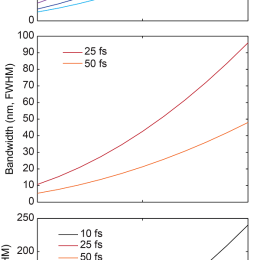
Phase retrieval
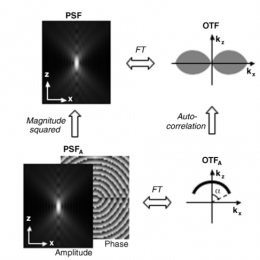
Microscopists are often adapting techniques pioneered for astronomy. Adaptive optics is the example you all probably know. Relatedly, phase retrieval was originally developed for telescopic imaging systems, and was adapted for high NA microscopic imaging systems…